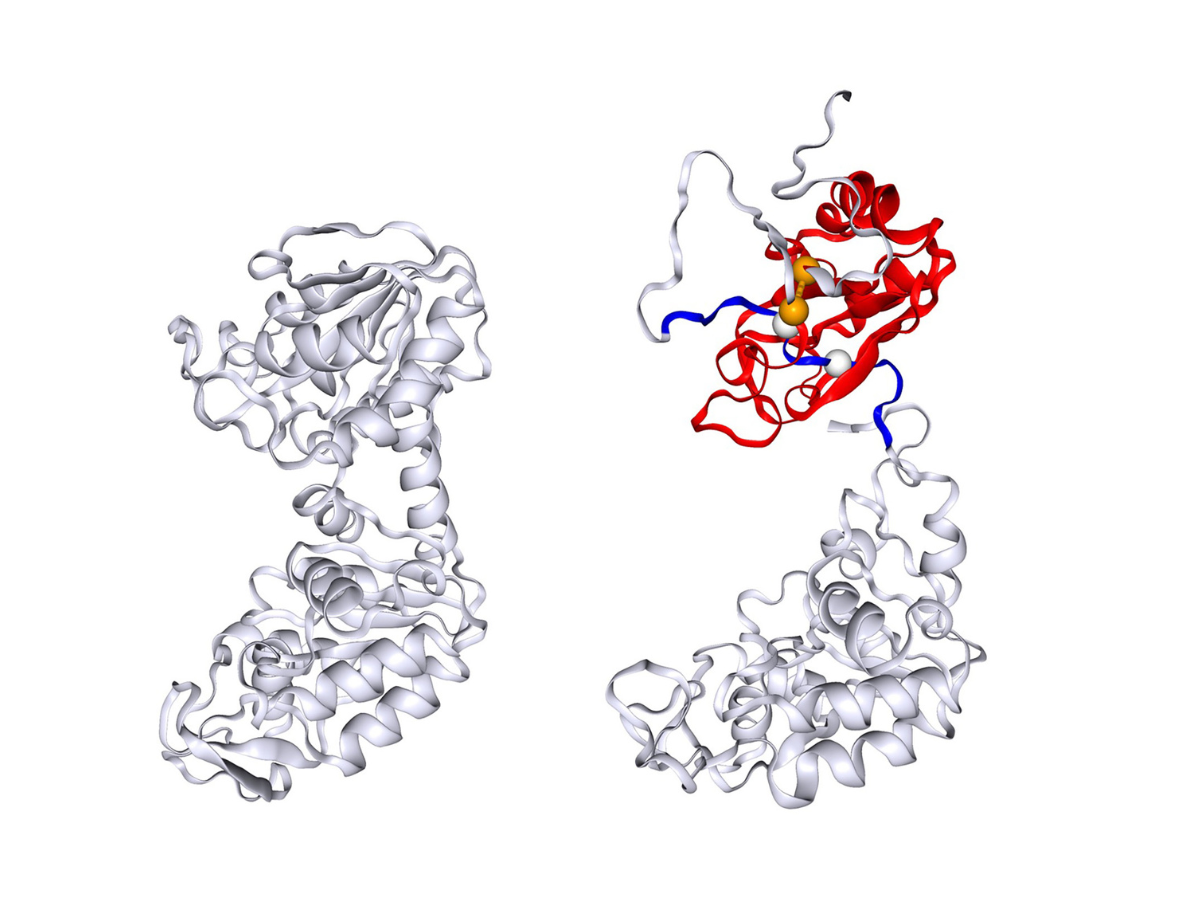
The image shows the native folded structure of the protein phosphoglycerate kinase (PGK) on the left and one of the misfolded PGK structures predicted in this study on the right, with the entangled regions highlighted in red and blue. Credit: Provided by Yang Jiang via Penn State News
ICDS co-hire leads research on protein misfolding
Posted on March 24, 2025EDITOR’S NOTE: A version of this story was originally published on Penn State News.
UNIVERSITY PARK, Pa. — A team of researchers at Penn State, including Ed O’Brien, Penn State Institute for Computational and Data Sciences co-hire and professor of chemistry in the Eberly College of Science, have found that proteins which naturally fold into complex structures to function correctly, can misfold and potentially lead to disease.
The new study, which was published on March 14 in the journal Science Advances, described a potential mechanism that could explain why some proteins misfold. According to the researchers, correcting this mishap requires high-energy or unfolding, which slows the folding process and leading to the first observed unexpected pattern in the 1990s.
“Misfolded proteins can malfunction and lead to disease. So, understanding the mechanisms involved in the folding processes can potentially help researchers prevent or develop treatments for diseases caused by misfolding,” O’Brien said in an article with Penn State News.
The team has identified this misfolding as a non-covalent lasso entanglement, which shows that a loop of the protein traps another segment of the protein, intertwining itself incorrectly, O’Brien said.
Researchers used a combination of computer simulations and refolding experiments to describe the folding kinetics of a protein called phosphoglycerate kinase (PGK). The folding pattern of PGK was observed for the first time experimentally over 25 years ago. PGK’s molecules followed a different pattern — or “stretched-exponential refolding kinetics — to the typical “two-state model” to reach a fully folded state. The structural mechanism to explain the difference in patterns was a mystery until the team’s study, which hypothesizes that this new class of misfolding could potentially be responsible for PGK’s pattern of folding.
The team built a computer model to simulate the folding process of the PGK protein and explored the intermediate stages of the folding processes to better understand the structure and see if there were changes that could explain the stretched refolding. The researchers found that the misfolded states predicted in the simulations were consistent with the structural signals observed in the refolded protein. Additionally, they suggested that these misfolded states were a crucial component of the observed stretched-exponential folding kinetics.
The research team includes: Yiang Jiang, assistant research professor of chemistry in the Eberly College of Science and first author on the paper; Ian Sitarik, Penn State graduate student; Hyebin Song, assistant professor of statistics at Penn State; Stephen Fried and his lab out of Johns Hopkins University; Yingzi Xia and Piyoosh Sharma, co-first authors at Johns Hopkins University.
Researchers used ICDS’ Roar supercomputer for their computer simulations and data analysis.
Share
Related Posts
- Professor receives NSF grant to model cell disorder in heart
- Featured Researcher: Nick Tusay
- Multi-institutional team to use AI to evaluate social, behavioral science claims
- NSF invests in cyberinfrastructure institute to harness cosmic data
- Center for Immersive Experiences set to debut, serving researchers and students
- Distant Suns, Distant Worlds
- CyberScience Seminar: Researcher to discuss how AI can help people avoid adverse drug interactions
- AI could offer warnings about serious side effects of drug-drug interactions
- Taking RTKI drugs during radiotherapy may not aid survival, worsens side effects
- Cost-effective cloud research computing options now available for researchers
- Costs of natural disasters are increasing at the high end
- Model helps choose wind farm locations, predicts output
- Virus may jump species through ‘rock-and-roll’ motion with receptors
- Researchers seek to revolutionize catalyst design with machine learning
- Resilient Resumes team places third in Nittany AI Challenge
- ‘AI in Action’: Machine learning may help scientists explore deep sleep
- Clickbait Secrets Exposed! Humans and AI team up to improve clickbait detection
- Focusing computational power for more accurate, efficient weather forecasts
- How many Earth-like planets are around sun-like stars?
- SMH! Brains trained on e-devices may struggle to understand scientific info
- Whole genome sequencing may help officials get a handle on disease outbreaks
- New tool could reduce security analysts’ workloads by automating data triage
- Careful analysis of volcano’s plumbing system may give tips on pending eruptions
- Reducing farm greenhouse gas emissions may plant the seed for a cooler planet
- Using artificial intelligence to detect discrimination
- Four ways scholars say we can cut the chances of nasty satellite data surprises
- Game theory shows why stigmatization may not make sense in modern society
- Older adults can serve communities as engines of everyday innovation
- Pig-Pen effect: Mixing skin oil and ozone can produce a personal pollution cloud
- Researchers find genes that could help create more resilient chickens
- Despite dire predictions, levels of social support remain steady in the U.S.
- For many, friends and family, not doctors, serve as a gateway to opioid misuse
- New algorithm may help people store more pictures, share videos faster
- Head named for Ken and Mary Alice Lindquist Department of Nuclear Engineering
- Scientific evidence boosts action for activists, decreases action for scientists
- People explore options, then selectively represent good options to make difficult decisions
- Map reveals that lynching extended far beyond the deep South
- Gravitational forces in protoplanetary disks push super-Earths close to stars
- Supercomputer cluster donation helps turn high school class into climate science research lab
- Believing machines can out-do people may fuel acceptance of self-driving cars
- People more likely to trust machines than humans with their private info
- IBM donates system to Penn State to advance AI research
- ICS Seed Grants to power projects that use AI, machine learning for common good
- Penn State Berks team advances to MVP Phase of Nittany AI Challenge
- Creepy computers or people partners? Working to make AI that enhances humanity
- Sky is clearing for using AI to probe weather variability
- ‘AI will see you now’: Panel to discuss the AI revolution in health and medicine
- Privacy law scholars must address potential for nasty satellite data surprises
- Researchers take aim at hackers trying to attack high-value AI models
- Girls, economically disadvantaged less likely to get parental urging to study computers
- Seed grants awarded to projects using Twitter data
- Researchers find features that shape mechanical force during protein synthesis
- A peek at living room decor suggests how decorations vary around the world
- Interactive websites may cause antismoking messages to backfire
- Changing how government assesses risk may ease fallout from extreme financial events
- Penn State’s Leadership in AI Research
- ICS co-sponsors Health, Environment Seed Grant Program
- Symposium at U.S. Capitol seeks solutions to election security
