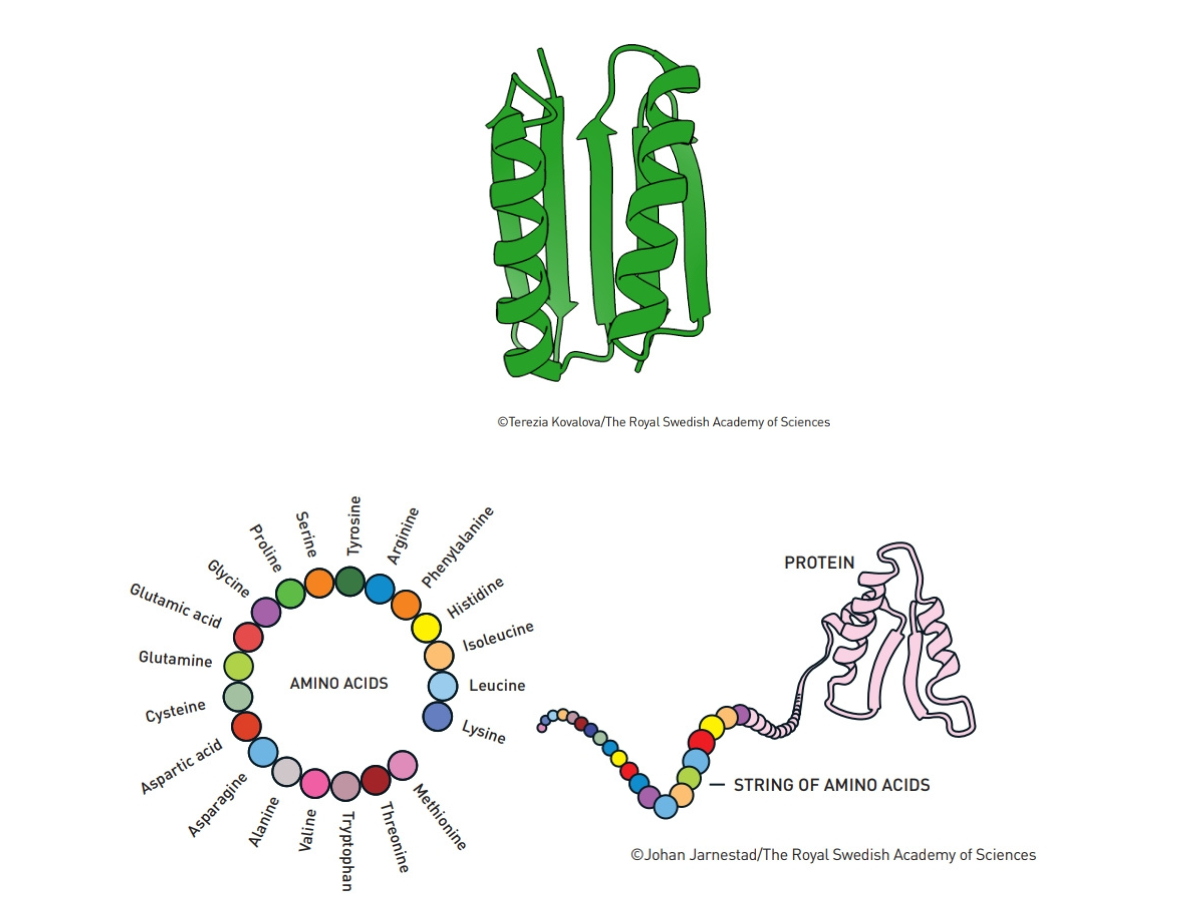
This illustration, at bottom, shows that a protein can consist of everything from tens of amino acids to several thousand amino acids. Credit: Johan Jarnestad/The Royal Swedish Academy of Sciences At top, this illustration shows in green, the first protein that was entirely different to all known existing proteins. Credit: Terezia Kovalova/The Royal Swedish Academy of Science
AlphaFold now more accessible for researchers
Posted on December 2, 2024The ability to infer the 3D structure of a protein knowing only its amino acid composition has been considered the holy grail of structural biology for decades. Advances in applied machine learning and artificial intelligence have resulted in a breakthrough — the AlphaFold AI software published by Google’s DeepMind AI team. This accomplishment was recently recognized with a Nobel Prize in Chemistry shared between David Baker of University of Washington (UW), and Demis Hassabis and John M. Jumper of Google DeepMind, London, UK. The most recent iteration of the algorithm (i.e., AlphaFold3) is now proprietary, obscuring technology that is poised to dramatically change the face of structural biology and the entire field of biomedical research. Vinay Saji Mathew, a doctoral student of Industrial and Manufacturing Engineering working with Dr. Soundar Kumara in the LISA Lab, has programmed an accessible platform in collaboration with researchers from the Institute for Computation and Data Sciences (ICDS) and Dr. William Lai’s lab at Cornell University.
This platform is available now as a service on the ICDS portal, ensuring AlphaFold is more accessible to researchers across diverse disciplines. Additionally, the group has made their platform code freely available and is collaborating with other national supercomputing centers to deploy AlphaFold as a supported service across the country. Vinay is pleased with the collaborative support by the ICDS because of the opportunities to network with researchers at other institutions and ability to access substantial GPU computing resources.
“Access to high-performance computing systems such as NCSA’s Delta and Jetstream2 provides the long-term computational power necessary for my doctoral research,” Mathew said.
The rapid adoption of the AlphaFold algorithm and its many open-source software variants is expected to continue across the broad domain of life sciences. A sizable number of nationally recognized national supercomputing sites (e.g., NIH BioWulf, UCSF Wynton, PSC, etc.) now offer AlphaFold2 to their user base, however adoption has been limited due to its complicated programmatic interface. This collaboration between Mathew, Gretta Kellogg at the Center for AI/ML to Industry (AIMI), Chad Bahrmann and Matt Hansen at ICDS, and Lai lab at Cornell University, has developed a web-portal interface for AlphaFold to the Penn State Community, with no coding experience required. Hansen, research and development engineer at ICDS, alongside the Research Innovations with Scientists and Engineers (RISE) team, has been instrumental in integrating this graphical interface into the ICDS Portal.
ICDS now has the resources to host and run AlphaFold and future upgrades to the platform include adding to the library of available structural algorithms (ESMFold, OpenFold, etc.) and adding the ability to design novel proteins from scratch (e.g., RFDiffusion). For more information about getting started with this platform, please send your request to icds@psu.edu.
To learn more about the code, click here.
To log into the Penn State ICDS Roar Collab Portal, click here.
The research project is generously funded by Cornell University BRC Epigenomics Core Facility (RRID:SCR_021287), Penn State Institute for Computational and Data Sciences (RRID:SCR_025154) and Penn State University Center for Applications of Artificial Intelligence and Machine Learning to Industry (AIMI) Core Facility (RRID:SCR_022867).
